Data Files#
The ccf_streamlines package uses several data files that contain the different flattened
representations, cortical streamlines, and other information.
To use many aspects of ccf_streamlines, these files must be downloaded to the user’s computer,
and their file paths are passed as parameters to various objects and functions.
This document links to those files and describes what they contain.
Streamlines#
File |
Description |
|---|---|
Contains the (linear) voxel locations of each streamline and a lookup table to find a streamline for each voxel. Warning: Large file (~0.5 GB) |
|
Lookup table of nearest surface voxel for cortical voxels, which are used to find the best-matching streamline. Warning: Large file (~200 MB) |
View lookups#
File |
Description |
Image |
|---|---|---|
Correspondence between view from back of isocortex and CCF volume. Can place hemispheres adjacent to each other with |
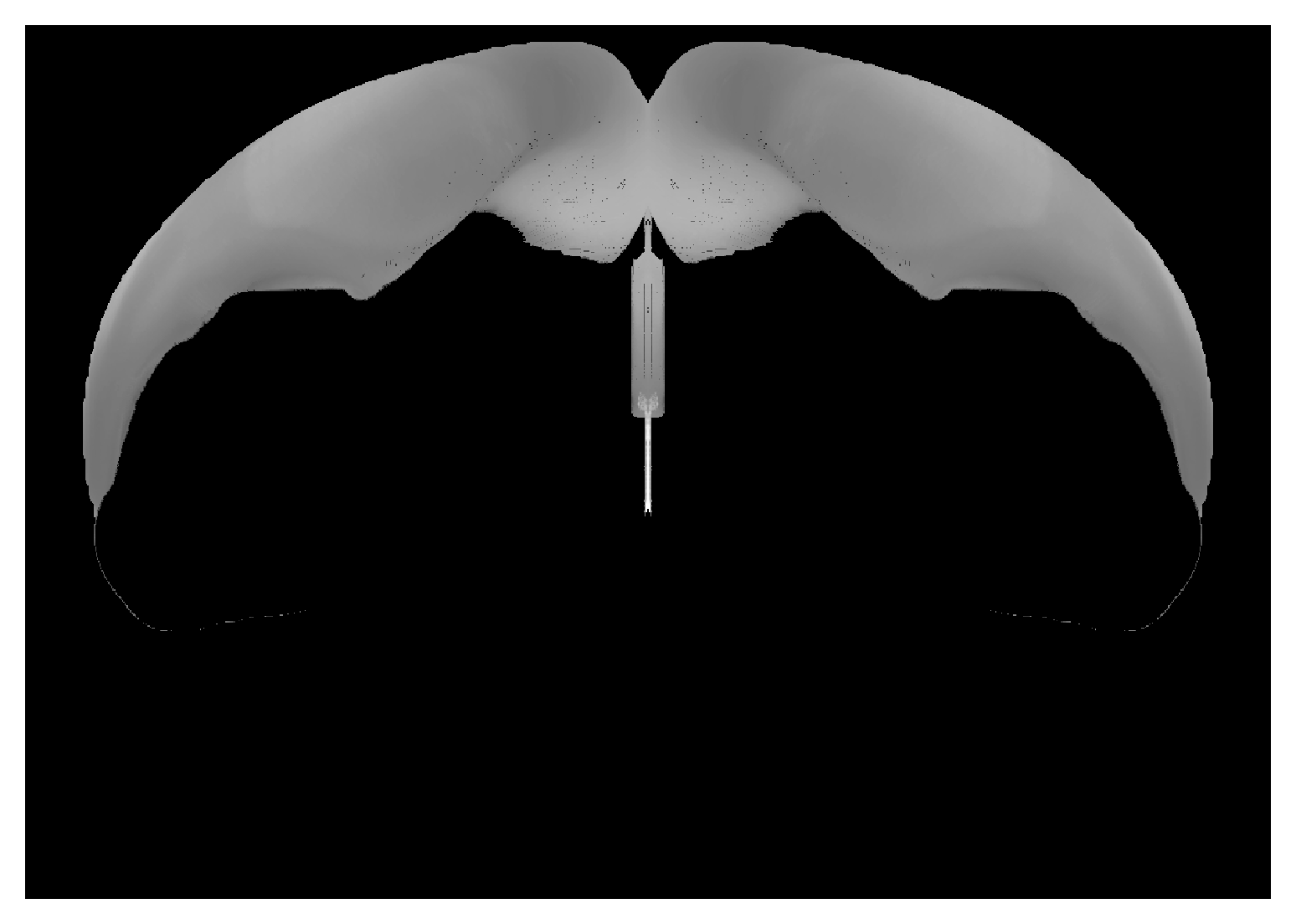
|
|
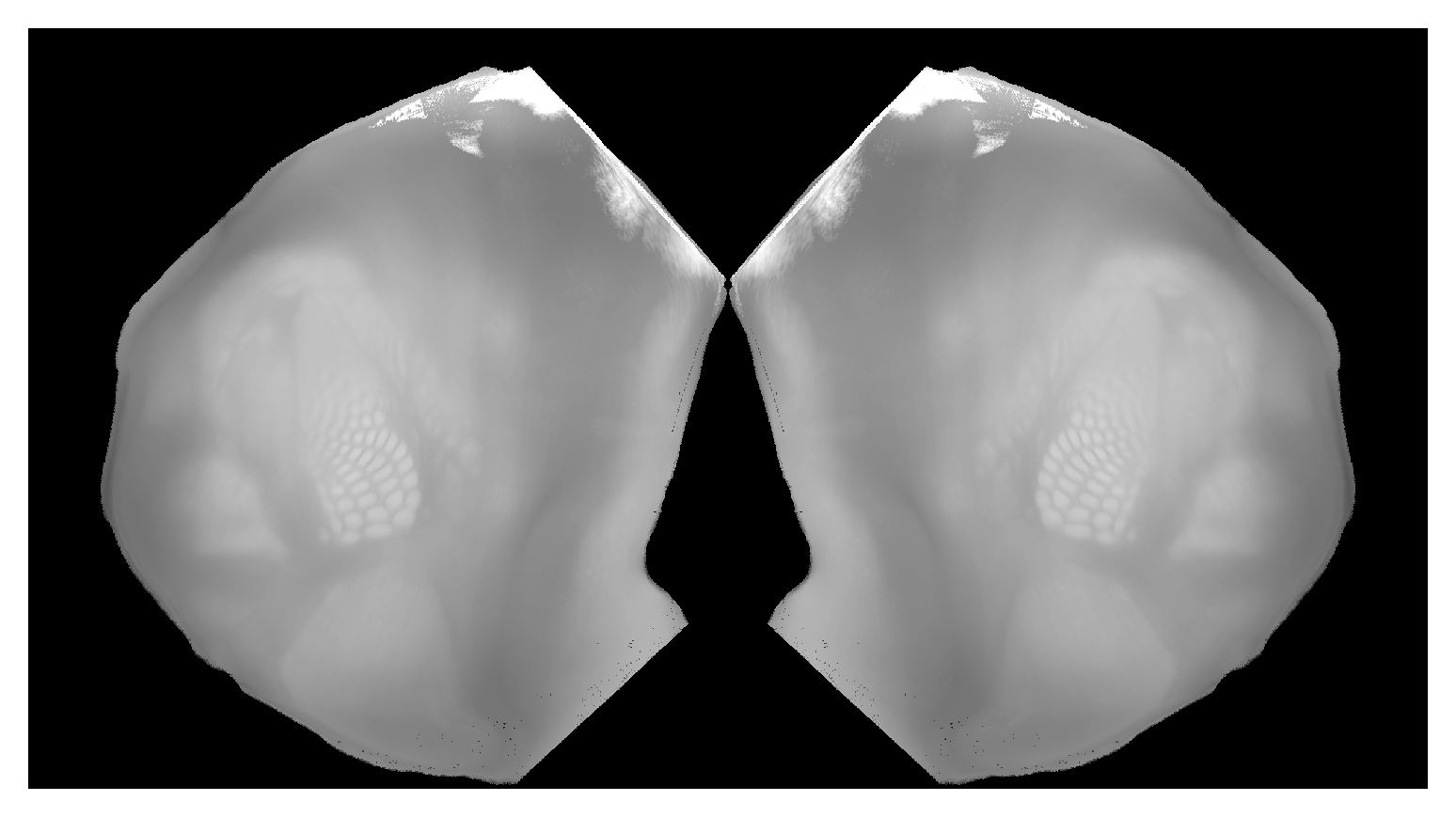
Correspondence between view from bottom of isocortex and CCF volume. Can place hemispheres adjacent to each other with |
|
|
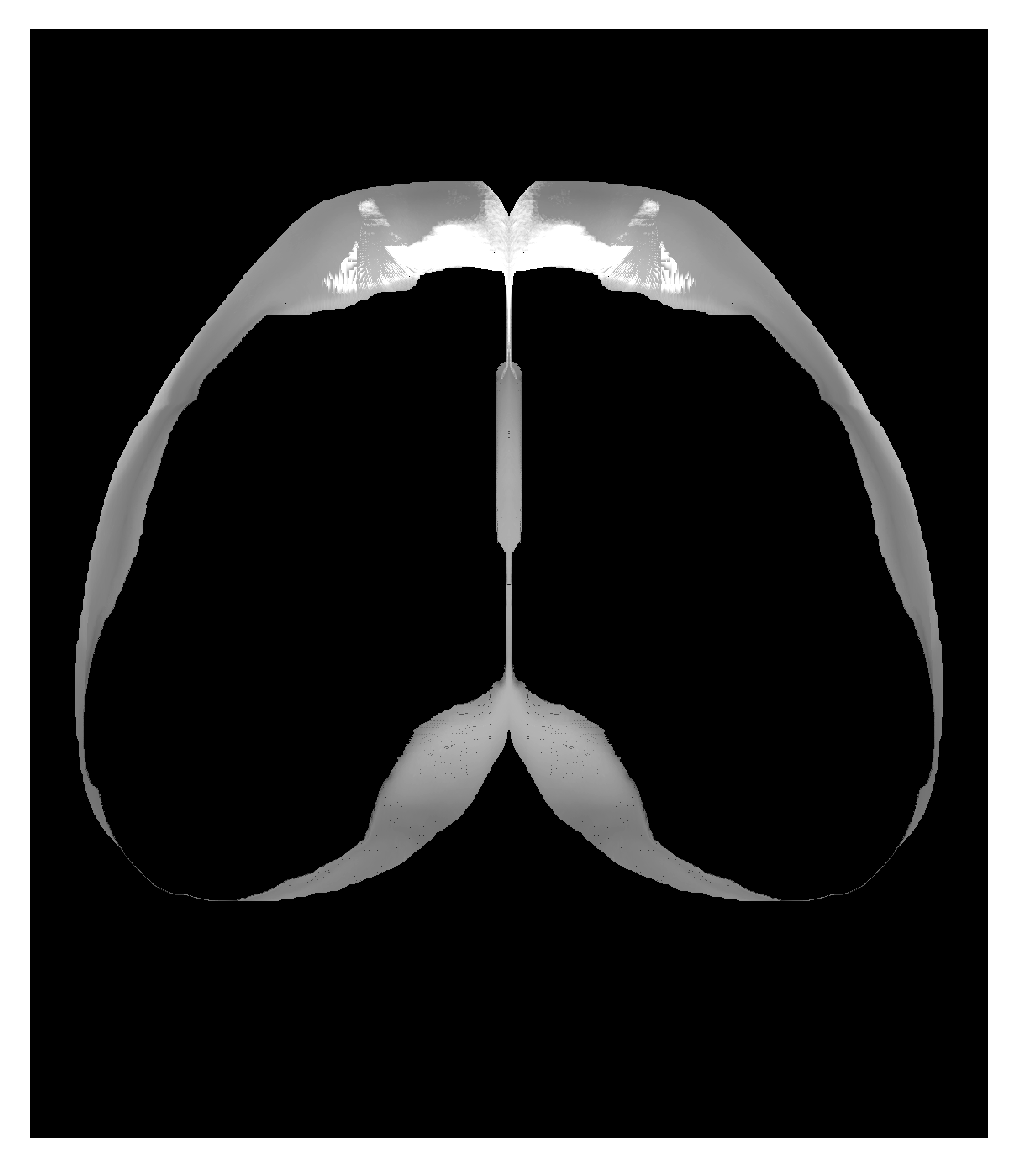
Correspondence between flattened map of all of isocortex and CCF volume - medial side has been adjusted so that the hemispheres are abutting at the center. Can place hemispheres adjacent to each other with |
|
|
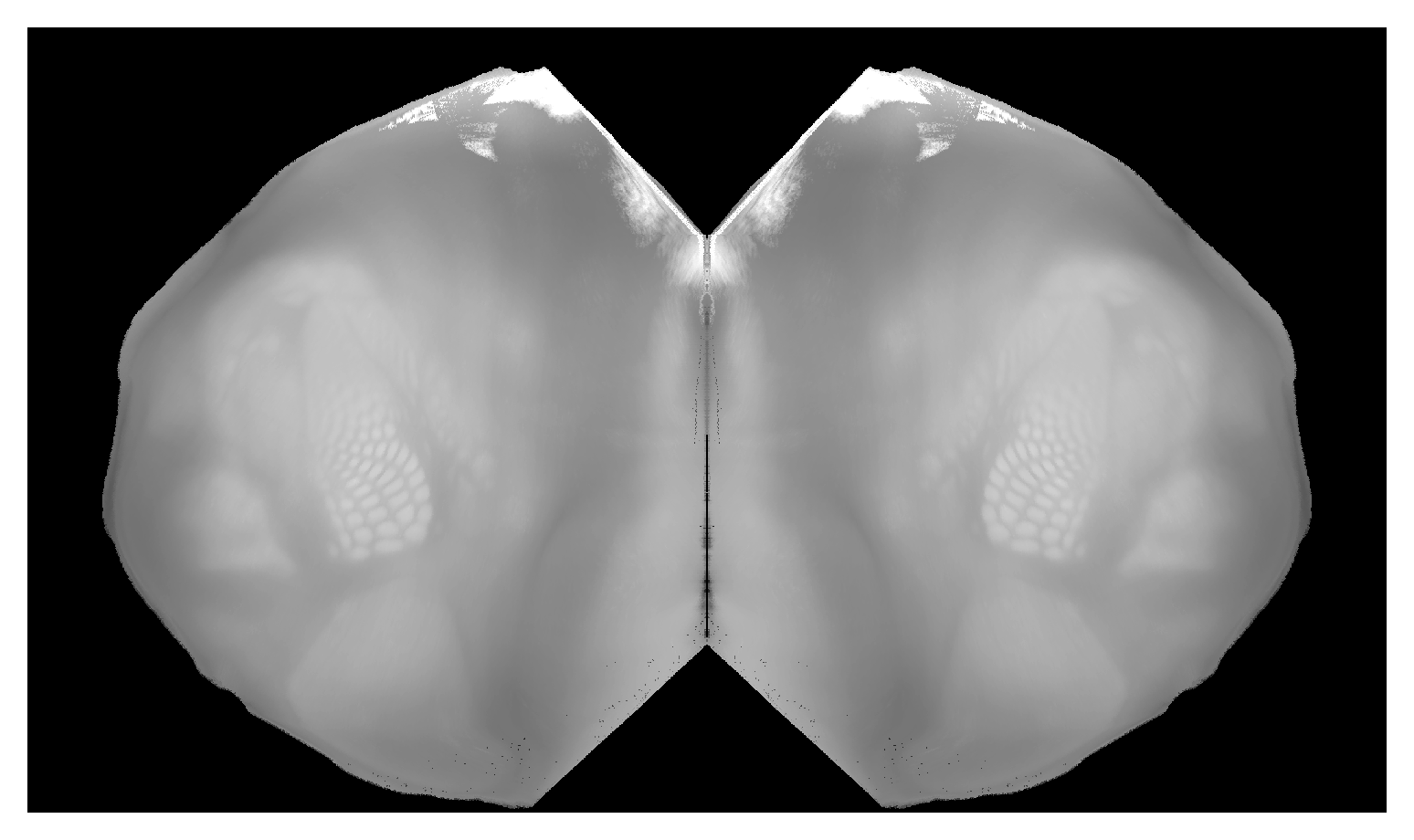
Correspondence between flattened map of all of isocortex and CCF volume. Can place hemispheres near each other with |
|
|
Correspondence between view from front of isocortex and CCF volume. Can place hemispheres adjacent to each other with |
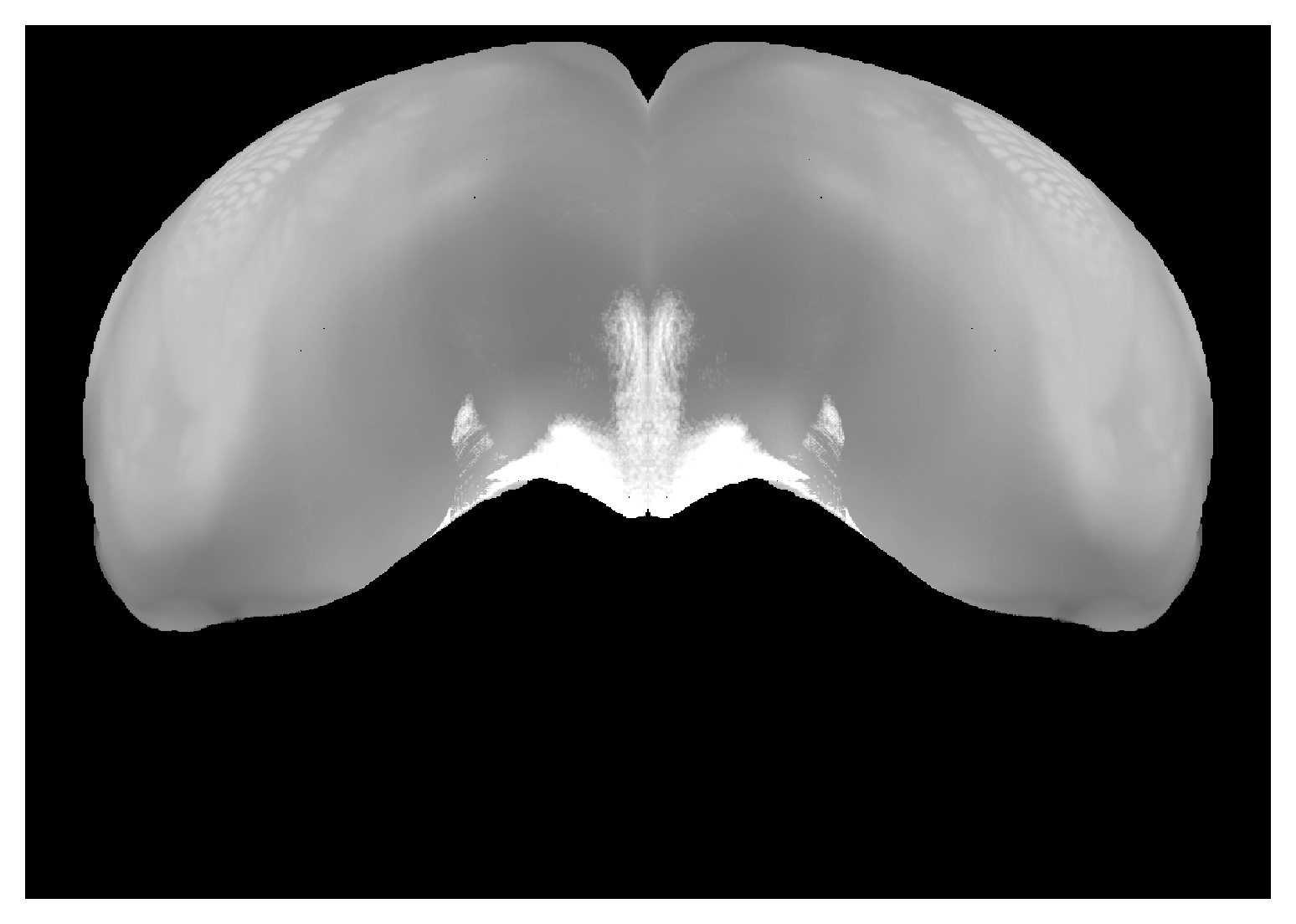
|
|
Correspondence between view from medial side of isocortex and CCF volume. Can place hemispheres near each other with |
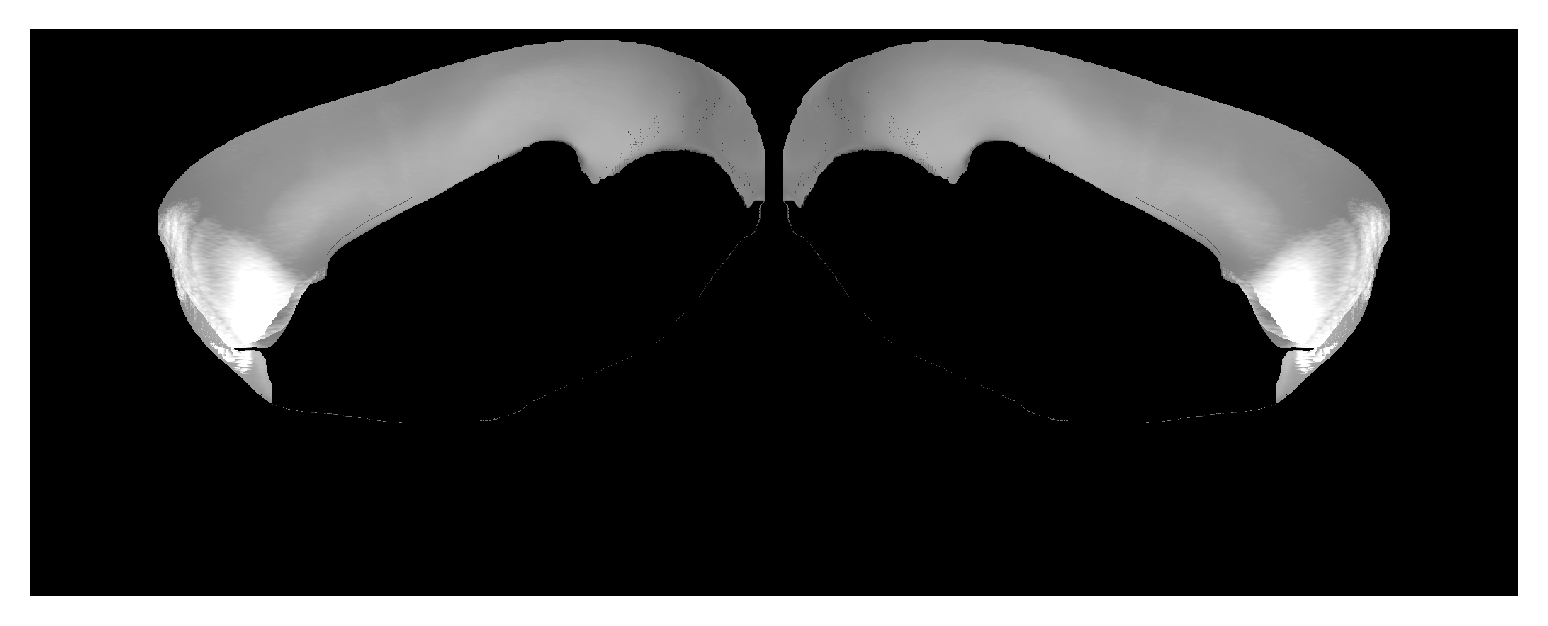
|
|
Correspondence between view from near the top but rotated to see more of lateral isocortex and CCF volume. Can place hemispheres near each other with |
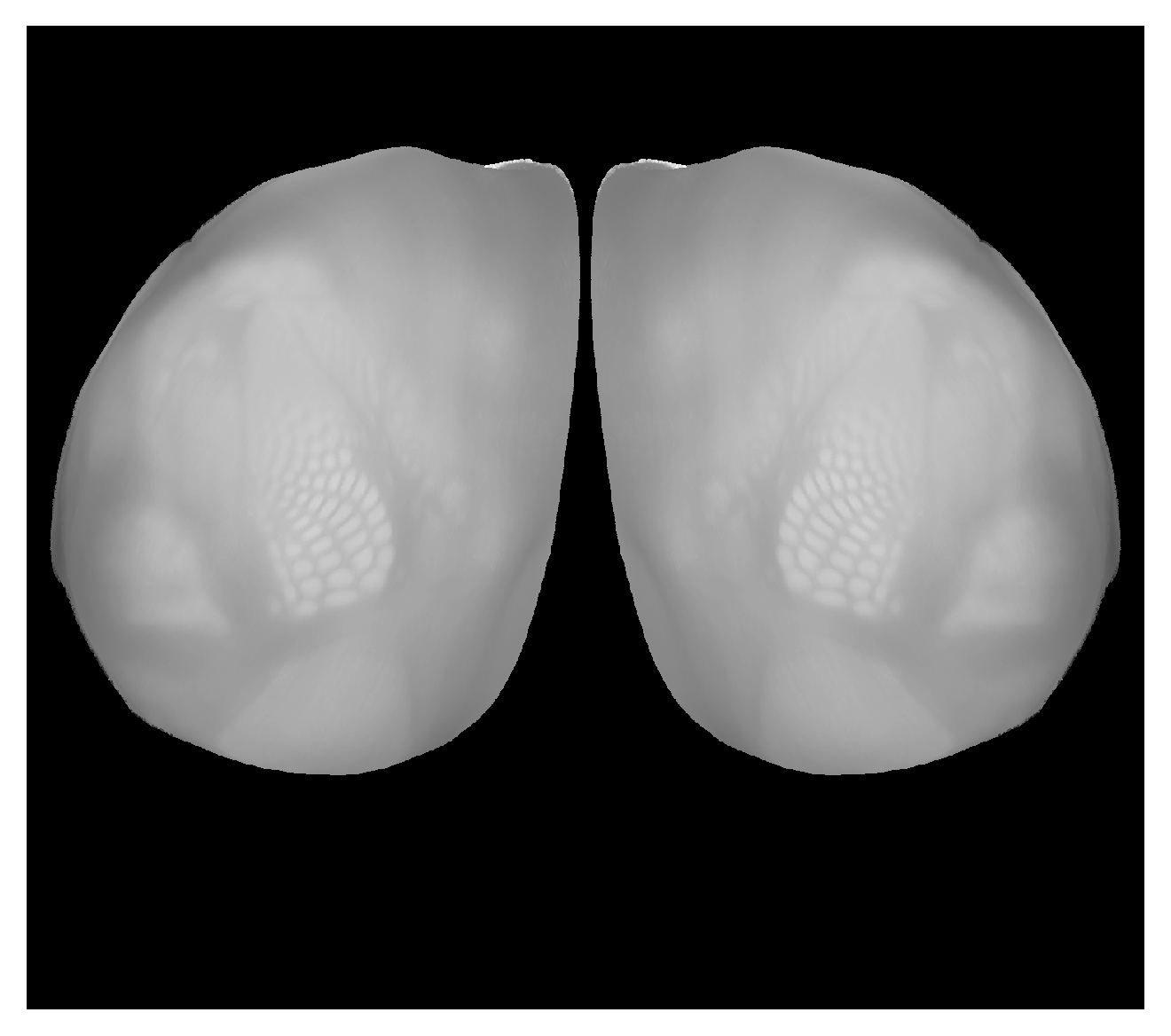
|
|
Correspondence between view from lateral side of isocortex and CCF volume. Can place hemispheres near each other with |
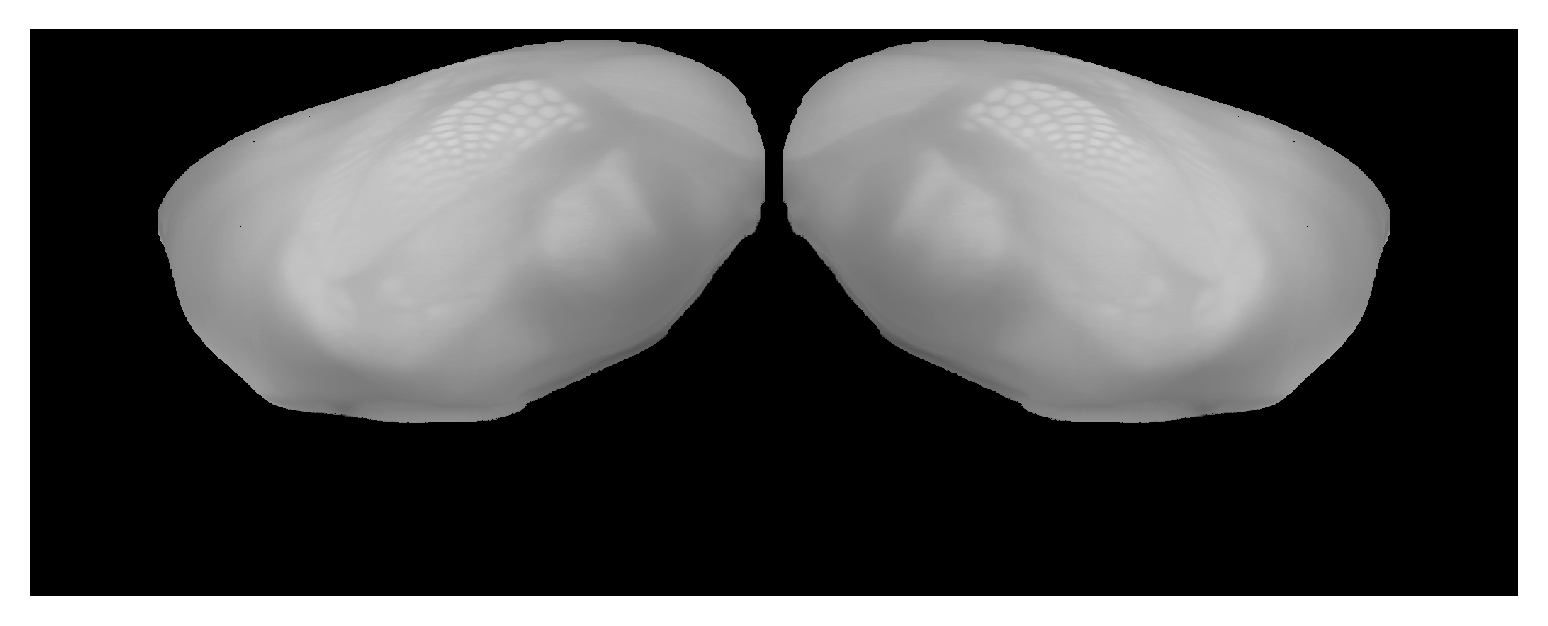
|
|
Correspondence between view from top of isocortex and CCF volume. Can place hemispheres adjacent to each other with |
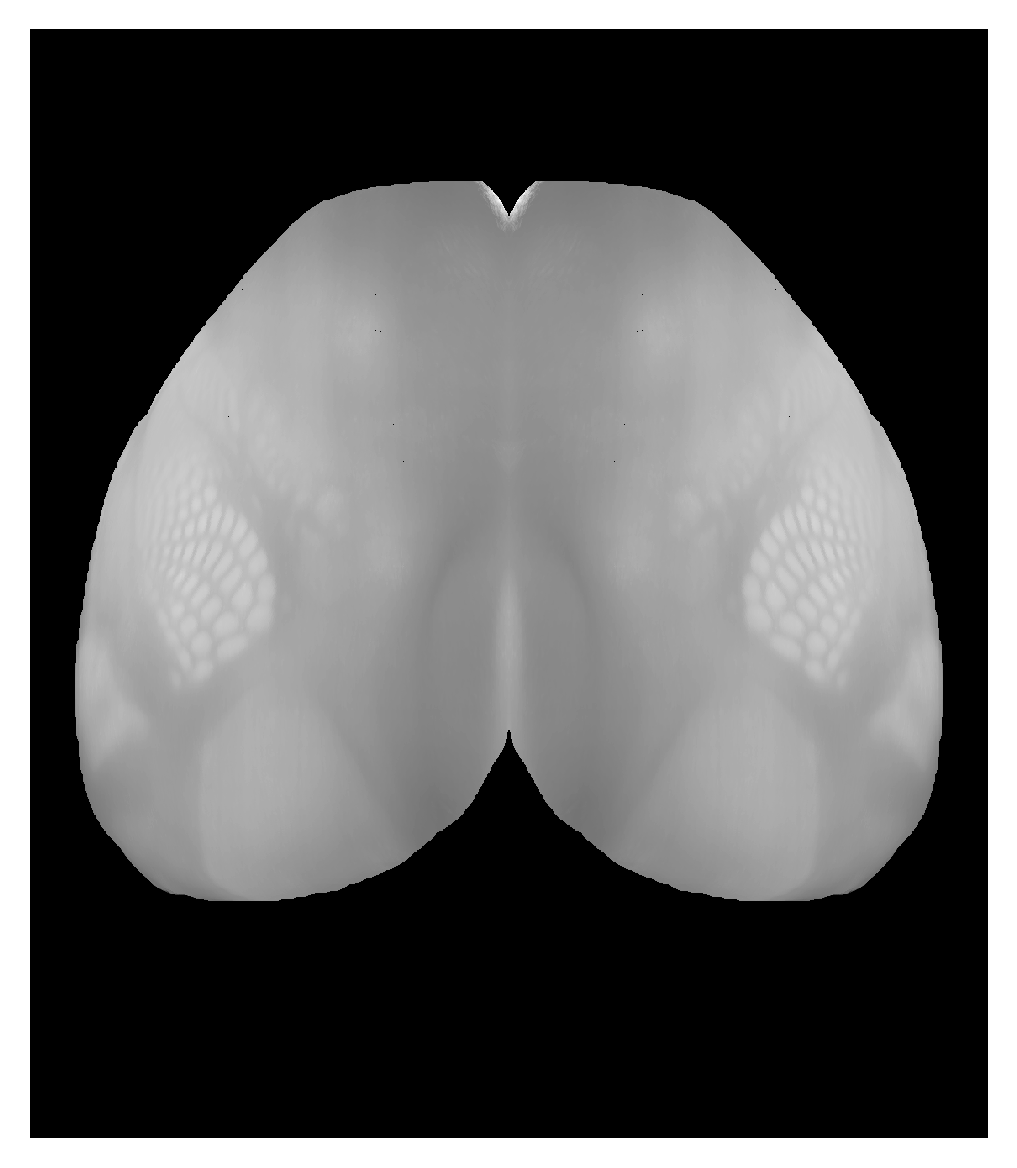
|
Atlas files (2D)#
File |
Description |
Image |
|---|---|---|
Atlas of view from back of isocortex |

|
|
Atlas of view from bottom of isocortex |
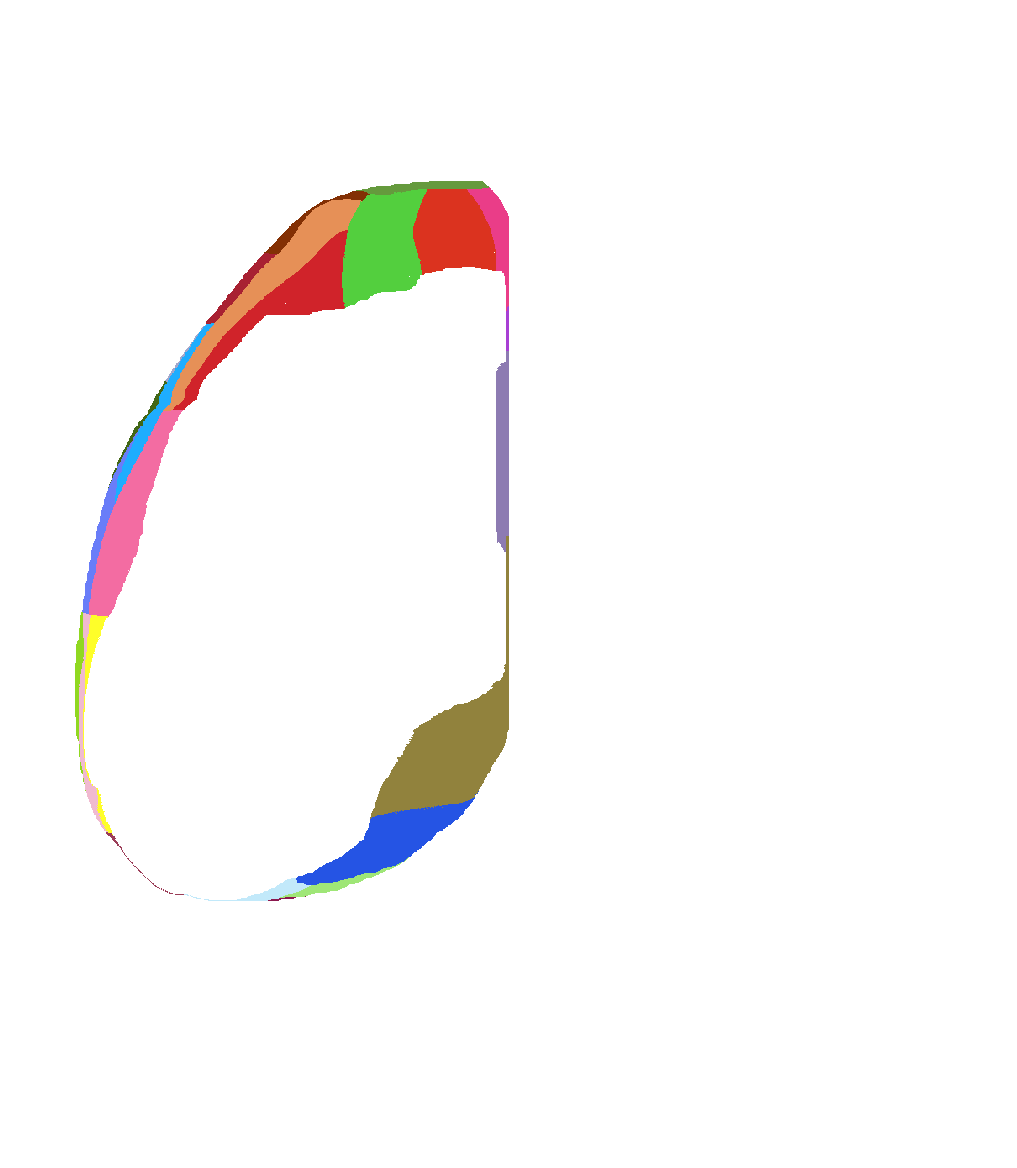
|
|
Atlas of flattened map of all of isocortex - medial side has been adjusted so that the hemispheres are abutting at the center. |
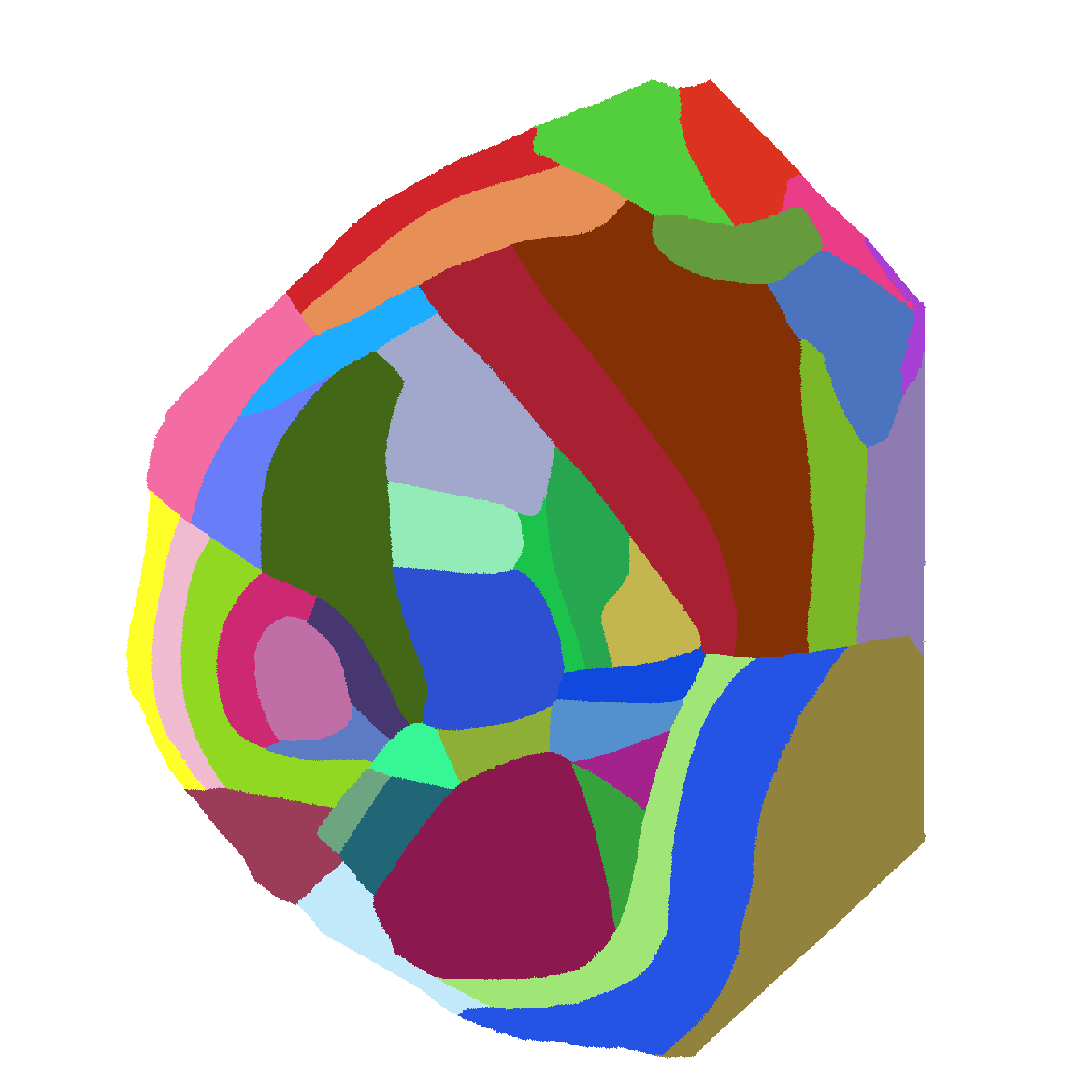
|
|
Atlas of flattened map of all of isocortex |
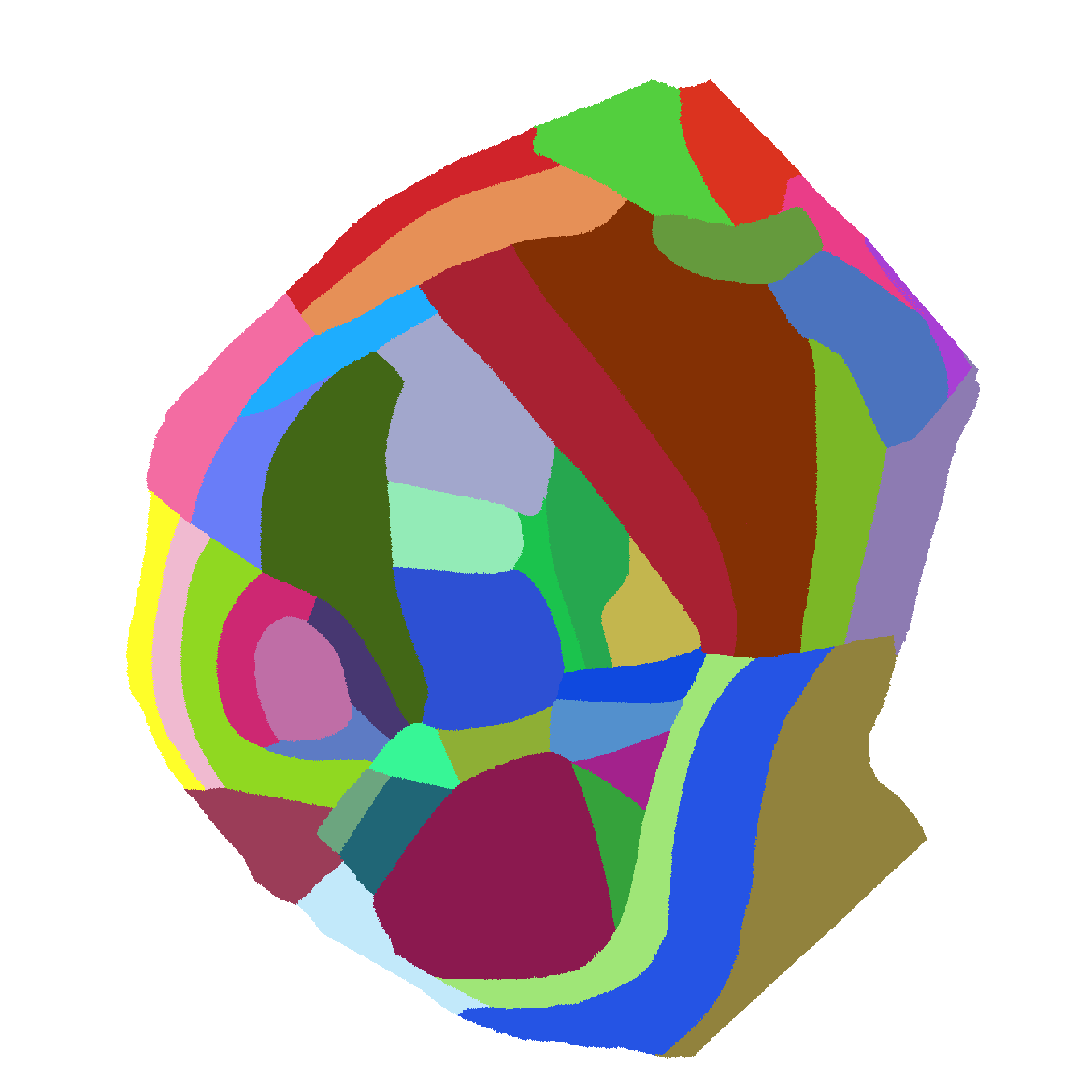
|
|
Atlas of view from front of isocortex |

|
|
Atlas of view from medial side of isocortex |
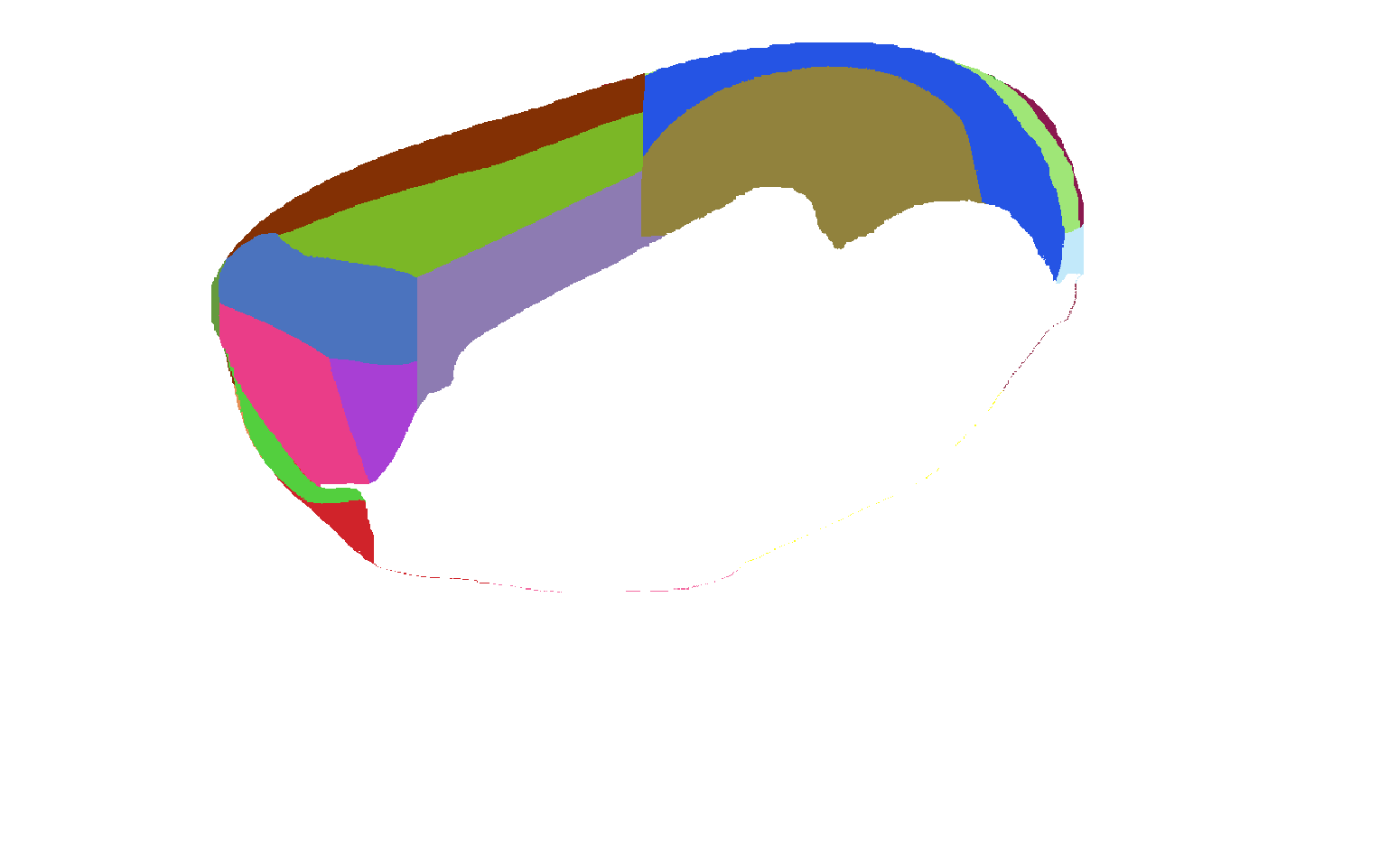
|
|
Atlas of view from near the top but rotated to see more of lateral isocortex |
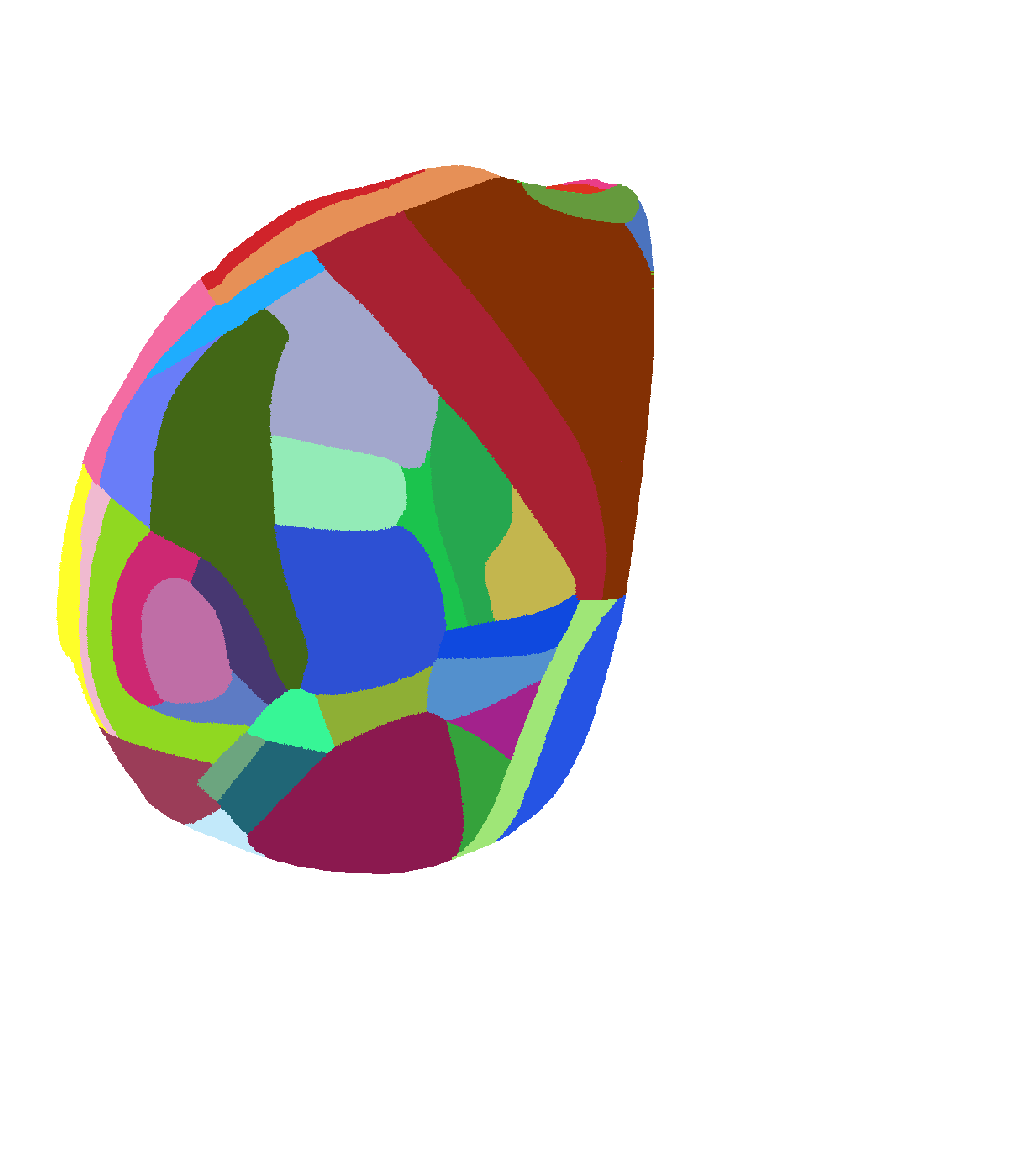
|
|
Atlas of view from lateral side of isocortex |
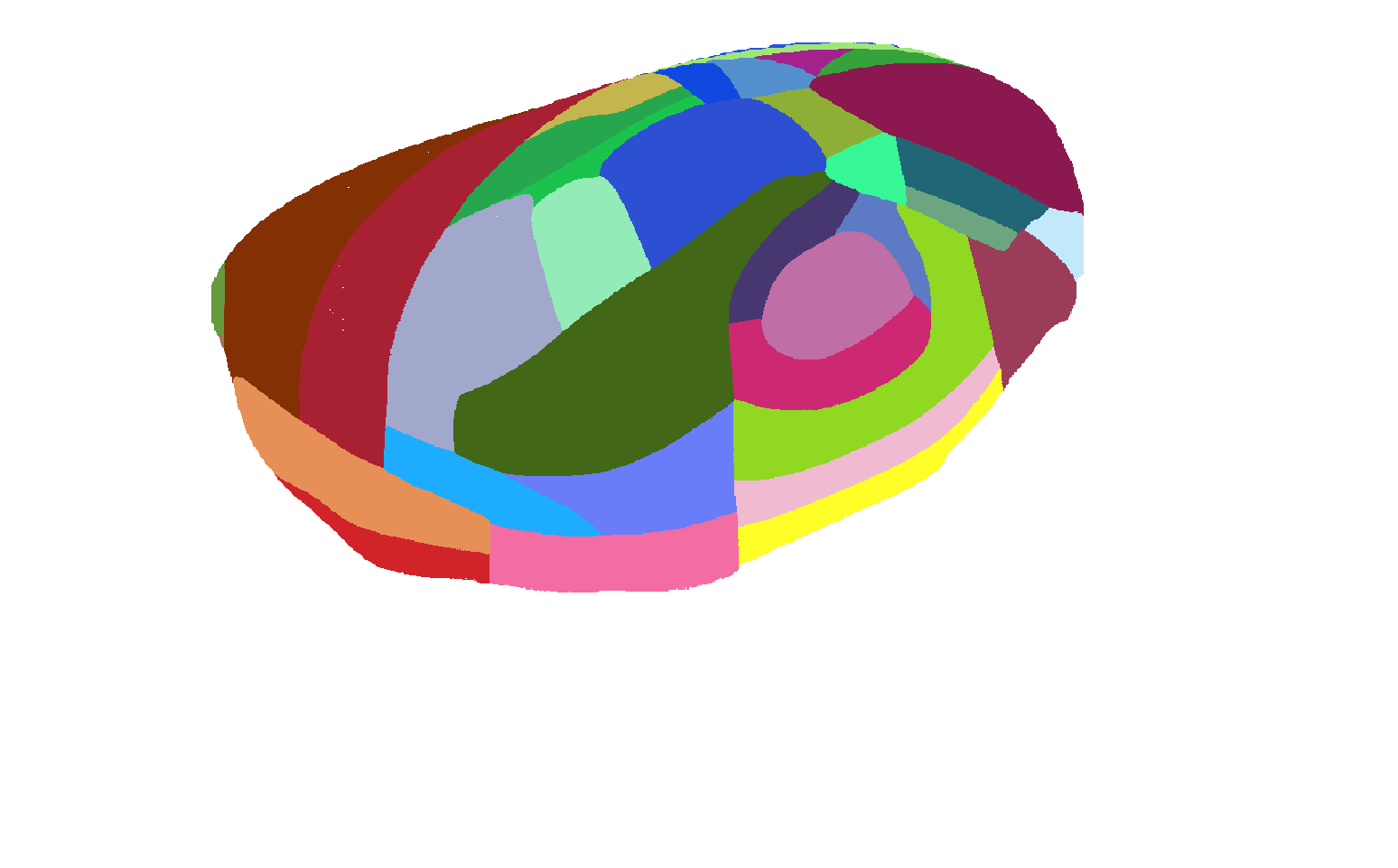
|
|
Atlas of view from top of isocortex |
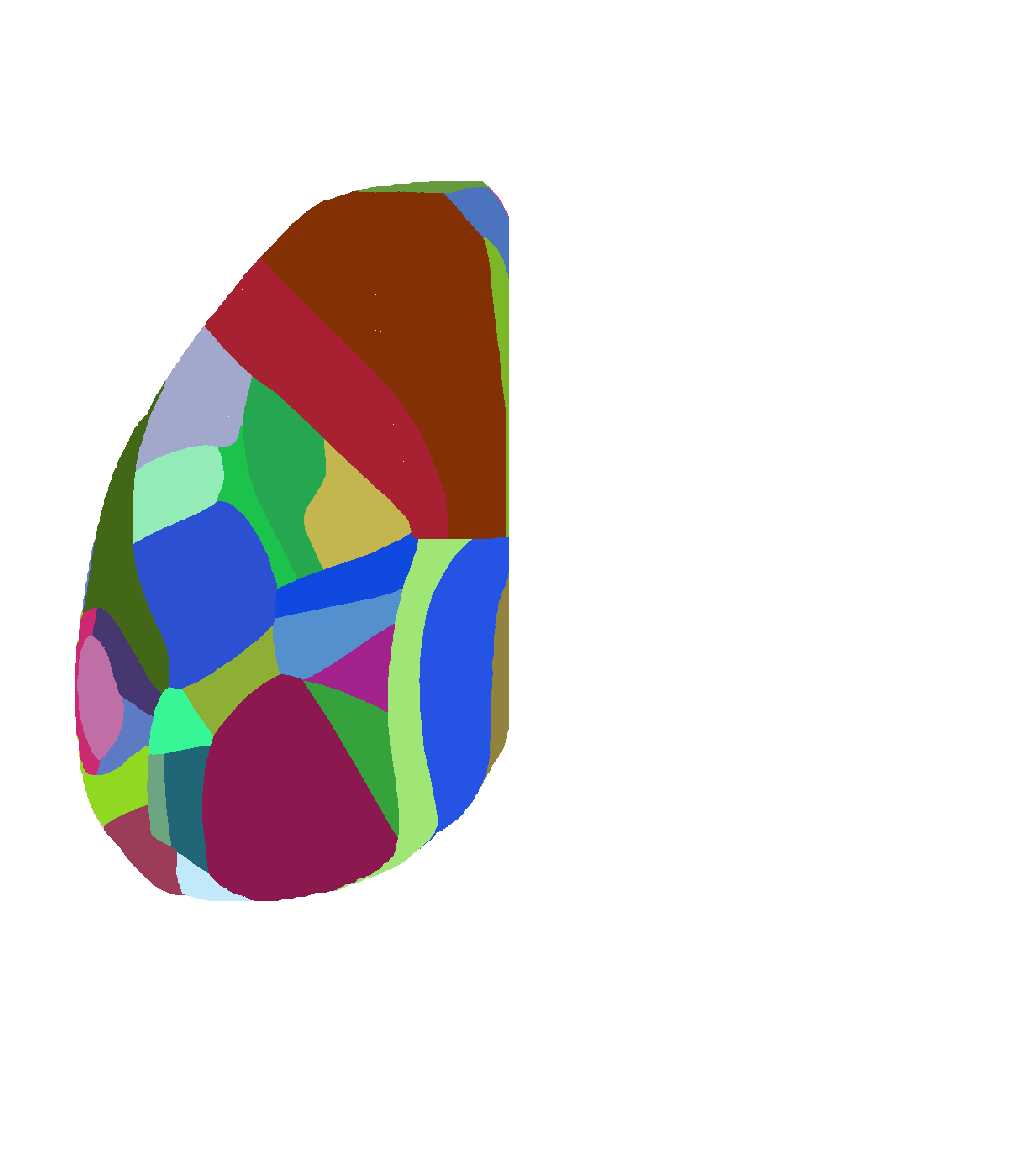
|
|
Metadata for cortical areas in atlas files. Contains identifiers, color, and CCF ontology acronyms. |
Isocortex metrics#
File |
Description |
|---|---|
A set of average layer depths to use as targets for normalizing layer thicknesses. Note: These were calculated from brain slices in mouse visual cortex - you may want to calculate or use your own thickness values instead. |
|
The starting depth, ending depth, and thickness of each layer for each streamline. Warning: Large file (~100 MB) |